This function allows you to calculate boundary/crossover point in logistic regression models.
cross_over(mod, cont_pred, grouping_var = FALSE, int_adj, slope_adj)Arguments
Examples
# Generate data
set.seed(1)
vot = rnorm(20, 15, 5)
vot = sort(vot)
phon1 = c(0,1,0,0,0,0,0,1,0,1,0,1,0,1,1,1,1,1,1,1)
group1 = rep('g1', 20)
df1 = data.frame(vot = vot, phon = phon1, group = group1)
phon2 = c(1,0,0,0,0,0,0,0,0,0,0,1,0,1,1,1,1,1,1,1)
group2 = rep('g2', 20)
df2 = data.frame(vot = vot, phon = phon2, group = group2)
df <- rbind(df1, df2)
# Fit model
glm1 <- glm(phon ~ vot, data = df, family = "binomial")
glm2 <- glm(phon ~ vot * group, data = df, family = "binomial")
# Get crossover point
cross_over(mod = glm1, cont_pred = 'vot')
#> [1] 16.20771
# Get crossover point of grouping variables
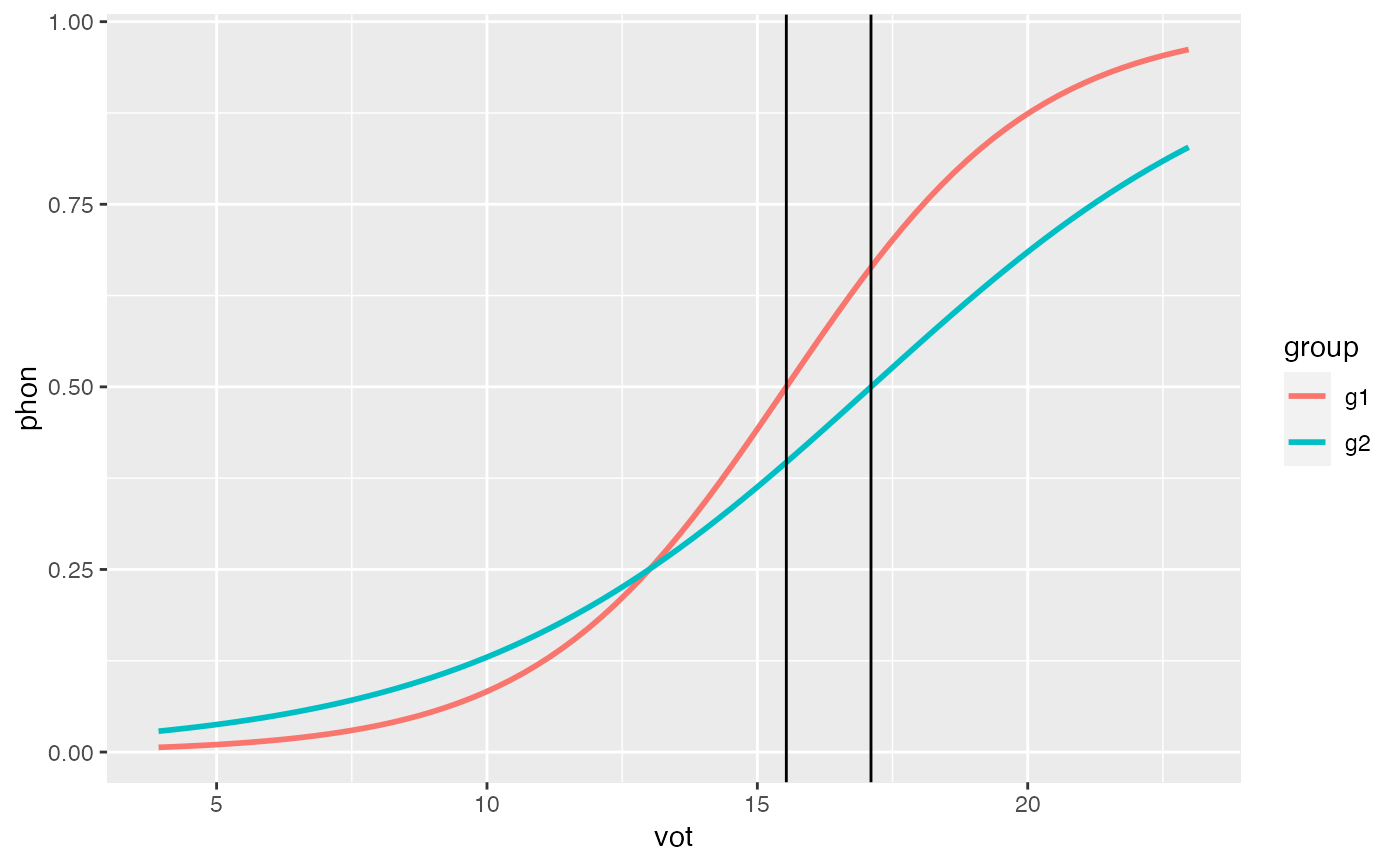
cross_over(mod = glm2, cont_pred = 'vot')
#> [1] 15.53595
cross_over(mod = glm2, cont_pred = 'vot', grouping_var = TRUE,
int_adj = 'groupg2', slope_adj = 'vot:groupg2')
#> [1] 17.10169
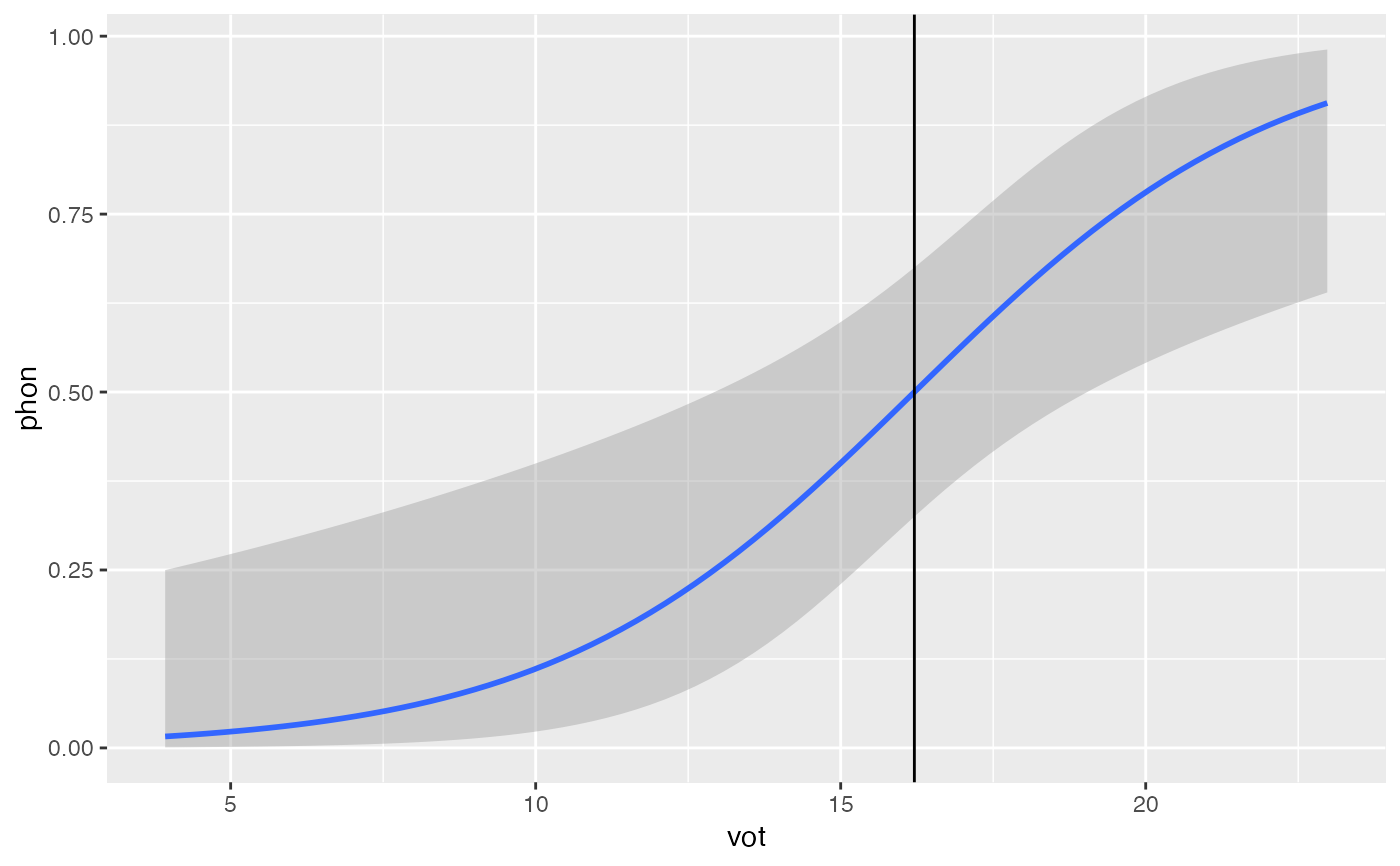
# Plot regression with crossover point
library(ggplot2)
ggplot(df, aes(x = vot, y = phon)) +
geom_smooth(method = 'glm', method.args = list(family = 'binomial')) +
geom_vline(xintercept = cross_over(mod = glm1, cont_pred = 'vot'))
#> `geom_smooth()` using formula = 'y ~ x'
 ggplot(df, aes(x = vot, y = phon, color = group)) +
geom_smooth(method = 'glm', method.args = list(family = 'binomial'),
se = FALSE) +
geom_vline(xintercept = cross_over(mod = glm2, cont_pred = 'vot')) +
geom_vline(xintercept = cross_over(mod = glm2, cont_pred = 'vot',
grouping_var = TRUE,
int_adj = 'groupg2',
slope_adj = 'vot:groupg2'))
#> `geom_smooth()` using formula = 'y ~ x'
ggplot(df, aes(x = vot, y = phon, color = group)) +
geom_smooth(method = 'glm', method.args = list(family = 'binomial'),
se = FALSE) +
geom_vline(xintercept = cross_over(mod = glm2, cont_pred = 'vot')) +
geom_vline(xintercept = cross_over(mod = glm2, cont_pred = 'vot',
grouping_var = TRUE,
int_adj = 'groupg2',
slope_adj = 'vot:groupg2'))
#> `geom_smooth()` using formula = 'y ~ x'